Experts in identifying SNPs for crop genetic improvement
SNP genotyping service for plant breeding
Single nucleotide polymorphisms (SNPs) are rapidly turning into the molecular marker of choice in crop plants because of their high level of polymorphism and reproducibility. In fact, the International Union for the Protection of New Varieties of Plants recommends SNPs and microsatellites as preferred markers for DNA fingerprinting and varietal characterisation (UPOV 2011).
Our SNP genotyping service is tailored to crop genetic improvement and plant breeding. At AllGenetics, we are able to identify thousands of SNPs among breeding lines, varieties, or cultivars of a crop species in an efficient and cost-effective manner. We sequence the genomes of the individuals under study to identify and call SNPs located within genes of interest. By doing so you receive a panel of SNPs which can be used for a number of applications, including:
- Cultivar or varietal fingerprinting.
- Verification of the varietal purity of seed lots.
- Protection of newly produced cultivars.
- Analysis of genetic diversity in breeding lines, cultivars, and wild populations.
- Marker assisted selection (MAS).
- Genome-wide association studies (GWAS).
- Linkage map construction.
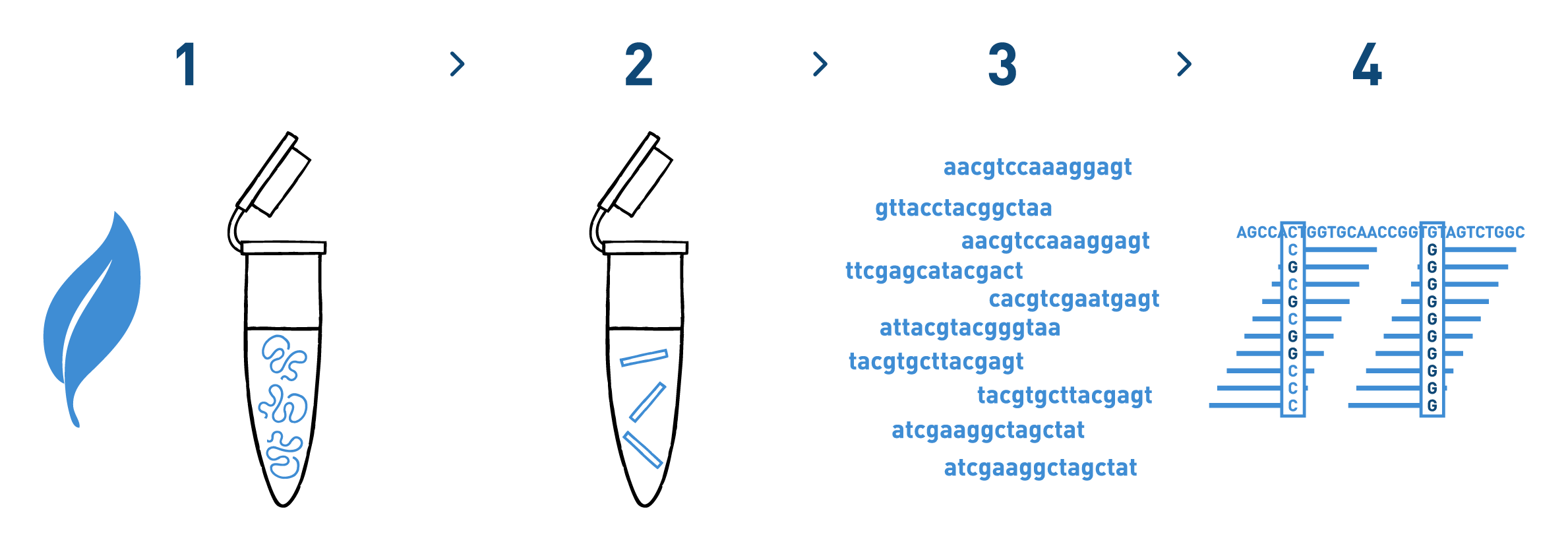
Step 1
We isolate DNA from your samples. We have adapted different DNA isolation protocols, depending on the starting biological material.
Step 2
We prepare genomic libraries using the kits recommended by Illumina.
Step 3
We sequence the libraries using the Illumina NovaSeq technology. Millions of reads per library are obtained in this step.
Step 4
We analyse the high-throughput sequencing data to identify and genotype the selected SNPs in each of the samples analysed.
What you receive
- A table with the alleles called at each locus in each individual.
- A list of gene-associated SNPs.
What we need
Your tissue samples appropriately preserved depending on the sample type (ethanol, silica gel, frozen, or another suitable preservation method). If required, we provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.