Experts in characterising microbial communities by DNA metabarcoding
Assessment of the impact of pollutants on the diversity of microbial communities
High-throughput sequencing (HTS) technologies have revolutionised the study of microbial communities, allowing the characterisation of species that cannot be detected by culture-dependent methods (around 99 % of total microbial diversity), and improving the taxonomic resolution provided by genetic fingerprinting techniques, such as DGGE and ARISA.
At AllGenetics, we use DNA metabarcoding to obtain diversity data for various taxonomic groups (bacteria, archaea, protists, fungi, nematodes) from a wide variety of environments, including soil, freshwater, and marine ecosystems.
With this approach, microbial ecotoxicology can now go beyond the use of a limited number of model microorganisms to investigations at the whole community level. This provides a more accurate assessment of the impacts of pollutants on microbial diversity, and the opportunity to search for better toxicity indicators for the entire microbial community.
In addition, a more rapid and cost-effective analysis allows for an increase in the number of monitored communities over space (e.g., pollution-impacted vs. non-polluted environments) and time (e.g., before and after a remediation treatment).
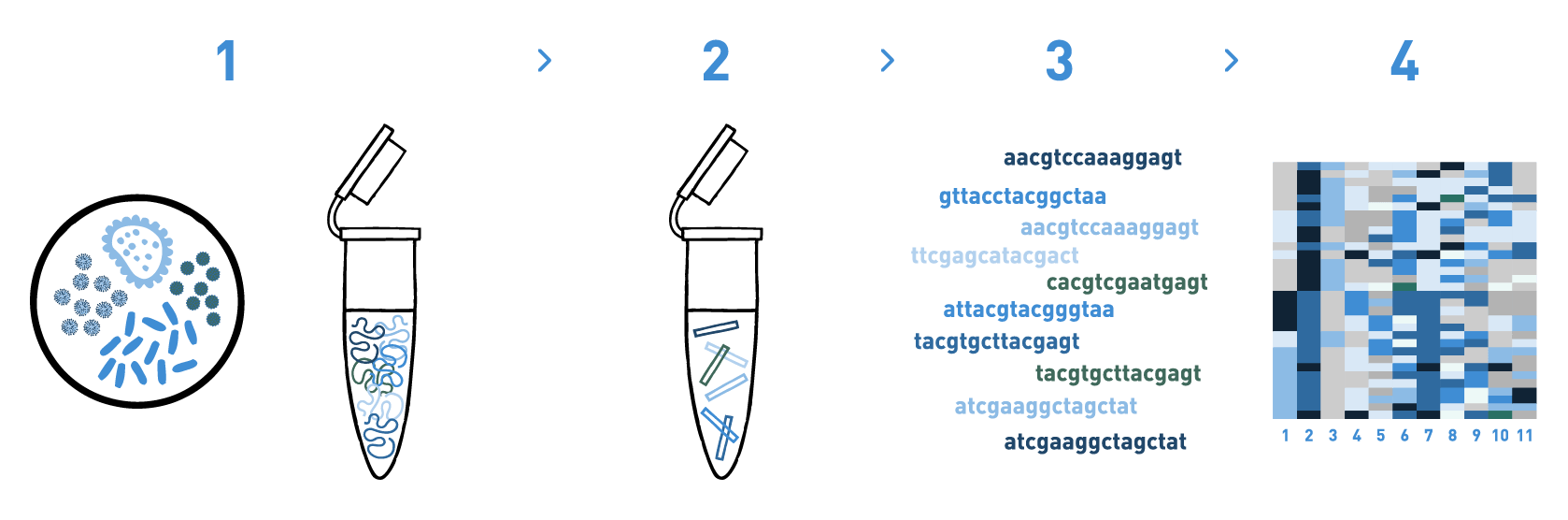
Step 1
We isolate DNA from different materials such as soil, water, sediments, etc.
Step 2
We prepare DNA metabarcoding libraries for the target taxonomic group(s) -bacteria, archaea, protists, fungi, and/or nematodes-. For this we use universal or in-house-developed primer pairs.
Step 3
We sequence the DNA metabarcoding libraries using Illumina's MiSeq or NovaSeq technologies.
Step 4
We analyse the data obtained in the previous step to get the taxonomic composition of the samples.
What you receive
- A list of the Operational Taxonomic Units (OTUs) retrieved per sample.
- The number of sequence reads assigned to each OTU in each sample.
- A graphic representation of the relative abundances of the different OTUs in each sample.
What we need
Your environmental samples which have been appropriately preserved (ethanol, silica gel, frozen, or another suitable preservation method). If required, we provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.