Experts in monitoring biodiversity by DNA metabarcoding
Get to know our biodiversity monitoring service
Traditional methods of species identification require each specimen to be processed individually. Consequently, they can be overly time-consuming and expensive for routine biomonitoring, which often involves the analysis of hundreds or thousands of specimens.
DNA metabarcoding now makes possible to sequence in parallel thousands of DNA barcodes from a bulk sample containing entire organisms (e.g., trap samples), or from DNA directly extracted from an environmental sample (environmental DNA or eDNA). Therefore, DNA metabarcoding infers the taxonomic composition of a complex sample without a prior step of specimen sorting.
With our biodiversity monitoring service, biodiversity surveys can move from individual specimen identification towards whole-community analysis, thereby yielding more comprehensive and resolute taxonomic inventories. Importantly, the direct analysis of environmental DNA allows for the detection of small-sized, rare, or elusive species, which, despite being traditionally absent from species censuses, may represent a significant fraction of the diversity in a biological community.
A more rapid and cost-effective analysis, in turn, enables an increase in the number of monitoring sites, which makes DNA metabarcoding an invaluable tool for long-term and large-scale environmental monitoring.
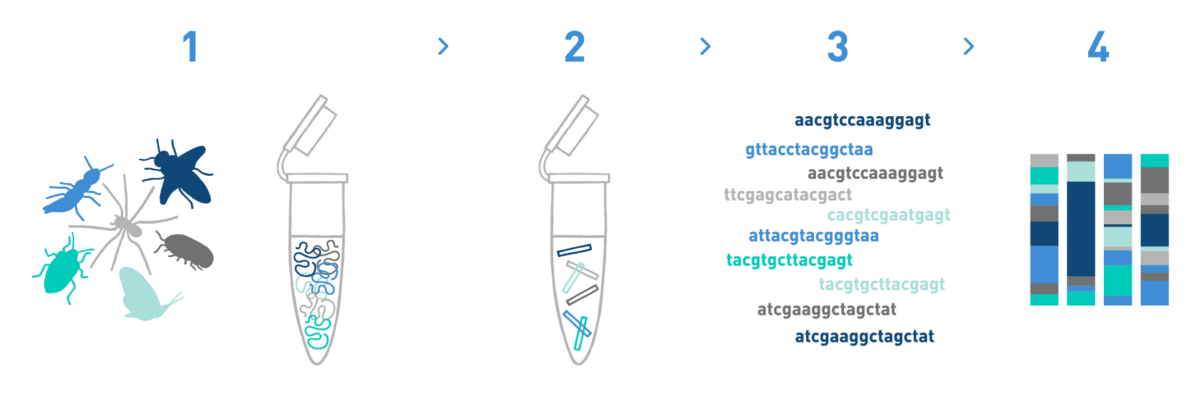
Step 1
We isolate DNA from different materials such as soil, water, sediments, etc.
Step 2
We prepare DNA metabarcoding libraries for the target taxonomic group(s) -bacteria, fungi, plants, protists, and/or metazoans-. For this we use universal or in-house-developed primer pairs.
Step 3
We sequence the DNA metabarcoding libraries using Illumina's MiSeq or NovaSeq technologies.
Step 4
We analyse the data obtained in the previous step to get the taxonomic composition of the samples.
What you receive
- A list of the Operational Taxonomic Units (OTUs) retrieved per sample.
- The number of sequence reads assigned to each OTU in each sample.
- A graphic representation of the relative abundances of the different OTUs in each sample.
What we need
Your environmental or tissue samples appropriately preserved depending on the sample type (ethanol, silica gel, frozen, or another suitable preservation method). If required, we provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.